Documentation
We developed BioKnExt, a Biological Knowledge Extractor, to automate the processing of literature and find variations in each gene. It allows a visualization of articles with highlighted entities and can be used to create or enrich generic databases.
BioKnExt website was developed with the Laravel Framework (www.laravel.com) to combine Information Retrieval (IR) and Named Entity Recignition (NER) approaches.
Below, some screenshots of possible searches with BioKnExt:
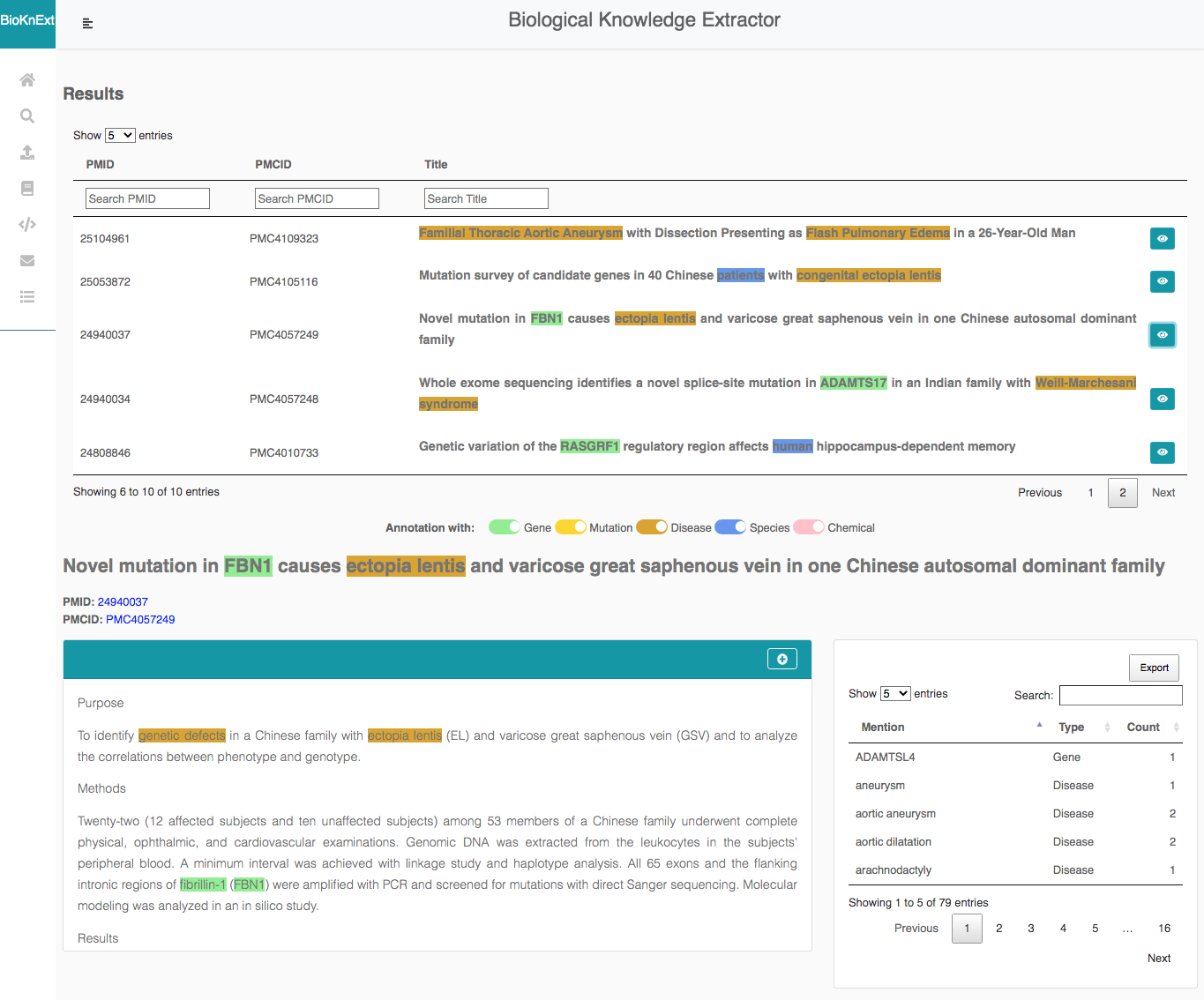
Users can query the system for a PMIDs list, a key word (disease, gene).
BioKnExt allows users to access the abstracts or the full texts of the request and to consult a summary table with the extracted entities and the context sentences. BioKnExt provides an intuitive visualization with colorful highlights and an easy navigation.
BioKnExt website was developed with the Laravel Framework (www.laravel.com) to combine Information Retrieval (IR) and Named Entity Recignition (NER) approaches.
Below, some screenshots of possible searches with BioKnExt:
Users can query the system for a PMIDs list, a key word (disease, gene).
BioKnExt allows users to access the abstracts or the full texts of the request and to consult a summary table with the extracted entities and the context sentences. BioKnExt provides an intuitive visualization with colorful highlights and an easy navigation.
PubTator Central provides a RESTful API that can be used to extract some entity types with different tools. These are state-of-the-art tools to extract entities such as genes/proteins, variations, chemicals, diseases, and species. We added nala to TmVar2.0 for the sequence variants extraction.
Tools used in BioKnExt:
| Tools | Help | Publications |
|---|---|---|
| TmVar2.0 | Is a text mining approach for extracting sequence variants from the literature. It can extract a wide range of variants described at the protein, DNA and RNA level, mentioned in abstracts or full texts. | TmVar TmVar2.0 |
| nala | Is a system for extracting sequence variants (genes and/or proteins) described in ST (standard) SST (semi-standard) or NL (natural language). This tool works on abstracts and full text and is not included in the PubTator Central tool. | nala |
| GNormPlus | Is a supervised approach to detect mentions of genes, gene families and protein domains from the text provided. | GNormPlus |
| TaggerOne | Is a tool based on a semi-Markovian structured linear classifier that can identify any type of entity (depending on the dataset it has been trained with). It is used here to identify disease mentions, chemicals and cell lines. | TaggerOne |
| SR4GN | Is a tool specialised in the recognition of species present in biomedical texts. It works with a recognition of the limits of the sentence in a first step and then detects the species names thanks to the Linnaeus dictionary. | SR4GN |
In order to facilitate the biocuration process, we have implemented a processing suite that allows users to submit a document in ".pdf", ".xlsx" or ".docx" format, transform it into a
format adapted to our process, and then process it with the TmVar2.0 and GNormPlus tools in order to extract the most likely gene-variation pairs. In addition to having access to the free literature
available via the main interface, the user can therefore provide the documents of his choice to extract information relating to the genes and variations of the latter. An API has been created for
the same purpose and allows the BioKnExt system to be queried to extract information from specific documents.